The improved software for predicting the tendency to form protein-peptide aggregates in living organisms was designed by researchers of University of Tehran
The researchers of the College of Science, University of Tehran succeeded in designing an improved software for predicting the propensity and ability to form protein-peptide aggregates in living organisms.
The researchers of the School of Biology in collaboration with the School of Mathematics, Statistics and Computer Sciences of the College of Sciences succeeded in designing and inventing a software for predicting the tendency of the formation of peptide and protein aggregates called SqFt.
This achievement has been achieved by Ms. Fatemeh Eshari, a researcher at the Protein Biotechnology Laboratory, under the guidance of Dr. Mehran Habibi Rezaei, a professor at the School of Biology, and Dr. Soudabeh Shemehsavar, an assistant professor at the School of Mathematics and Computer Science.
Dr. Habibi Rezaei, head of this research group, said about the importance of this achievement: In the conditions of oxidative stress, many natural proteins and peptides with known and sometimes unknown functions have different degrees of tendency to form amyloid accumulation.
As a result of the formation of amyloid accumulations, the normal function of proteins is lost and sometimes they become toxic. These conditions are observed in an age-dependent manner in diseases related to oxidative stress such as type 2 diabetes (T2DM), Alzheimer's (AD) and Parkinson's (PD), which are currently increasing in societies including our country.
He added: These amyloid accumulations are formed in Alzheimer's disease as extracellular amyloid plaques and coils of intracellular filaments, and in Parkinson's disease as intracellular deposits called Lewy bodies. Aggregation formation in proteins is a function of their physical chemistry and complex molecular structure. Therefore, providing the possibility of predicting the accumulation of proteins is important for adopting preventive strategies for amyloid-related diseases, as well as for the design and processing and functional evaluation of biological drugs in the specialized field of protein engineering.
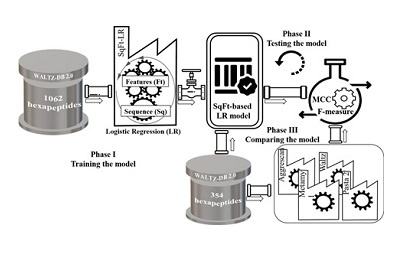
Dr. Habibi Rezaei emphasized: Until now, various tools and servers have been introduced to predict the accumulation of proteins and peptides; However, finding a model or server with higher sensitivity and accuracy based on understanding the effect of each of the intrinsic and structural features of peptides and proteins is still of interest to researchers. In order to achieve this achievement, first the direct effect of each of the characteristics in the aggregation process was investigated and a new model was presented to predict the accumulation of hexapeptides using the Machine Learning method.
Professor of School of Biology of University of Tehran added: WALTZ DB-2.0 server which consisted of 1416 amyloid and non-amyloid hexapeptides was used in order to design and test the model with the instructions of Dr. Soudabeh Shemehsavar, Associate Professor of Statistics. First, the model was designed using Logistic Regression and 354 hexapeptides were used to test the software and indicators and statistical analyzes were used to check the effectiveness of the model. In the following, the efficiency and reliability of the invented model was proved by comparing the desired calculation quantities with the values obtained from common and well-known servers such as Aggrescan, WALTZ, Metamyl and PASTA2.0.
The results of this research titled " Prediction of protein aggregation propensity employing SqFt-based logistic regression model" have recently been published in " International Journal of Biological Macromolecules " with an impact factor of 8.2.

Your Comment :